Upload Function Guide
Visualization Function Guide
Re-Visualization Function Guide
Result Files Download Guide
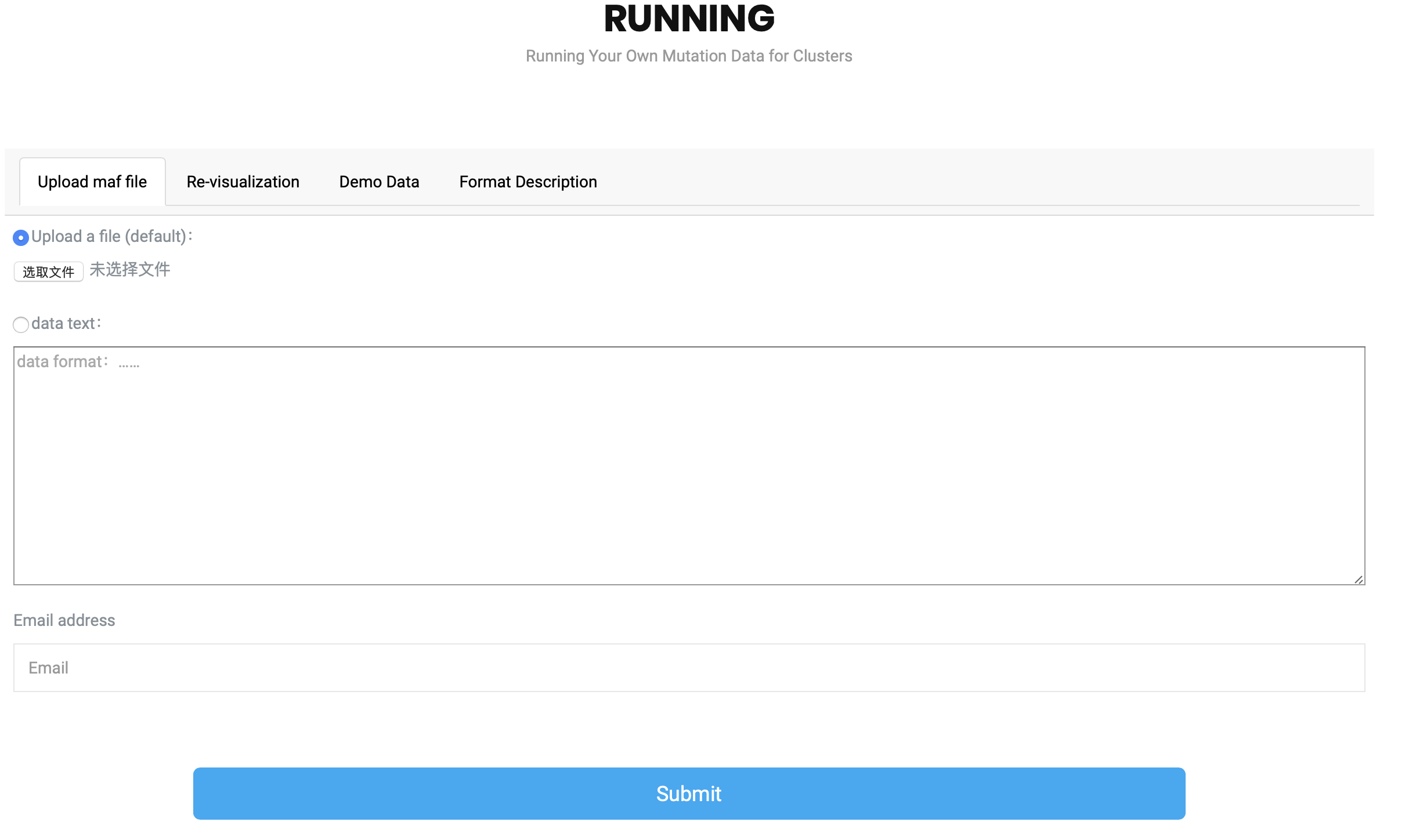
What is uploading ?
What format input file is required to run HotSpot3D?
The input file must contain 11 columns of data in the Format Description?
Can I change the column name?
If I haven't prepared the maf file yet, how should I use Hotspot3D?
Is the demo data consistent with the effect of running my own files?
Successfully running the software, but there has never been a result?
Must I waiting for result?
What is the meaning of the icons in different colors in the results page?
What is visualization?
How to choose protein structure?
What is the source of the protein structure?
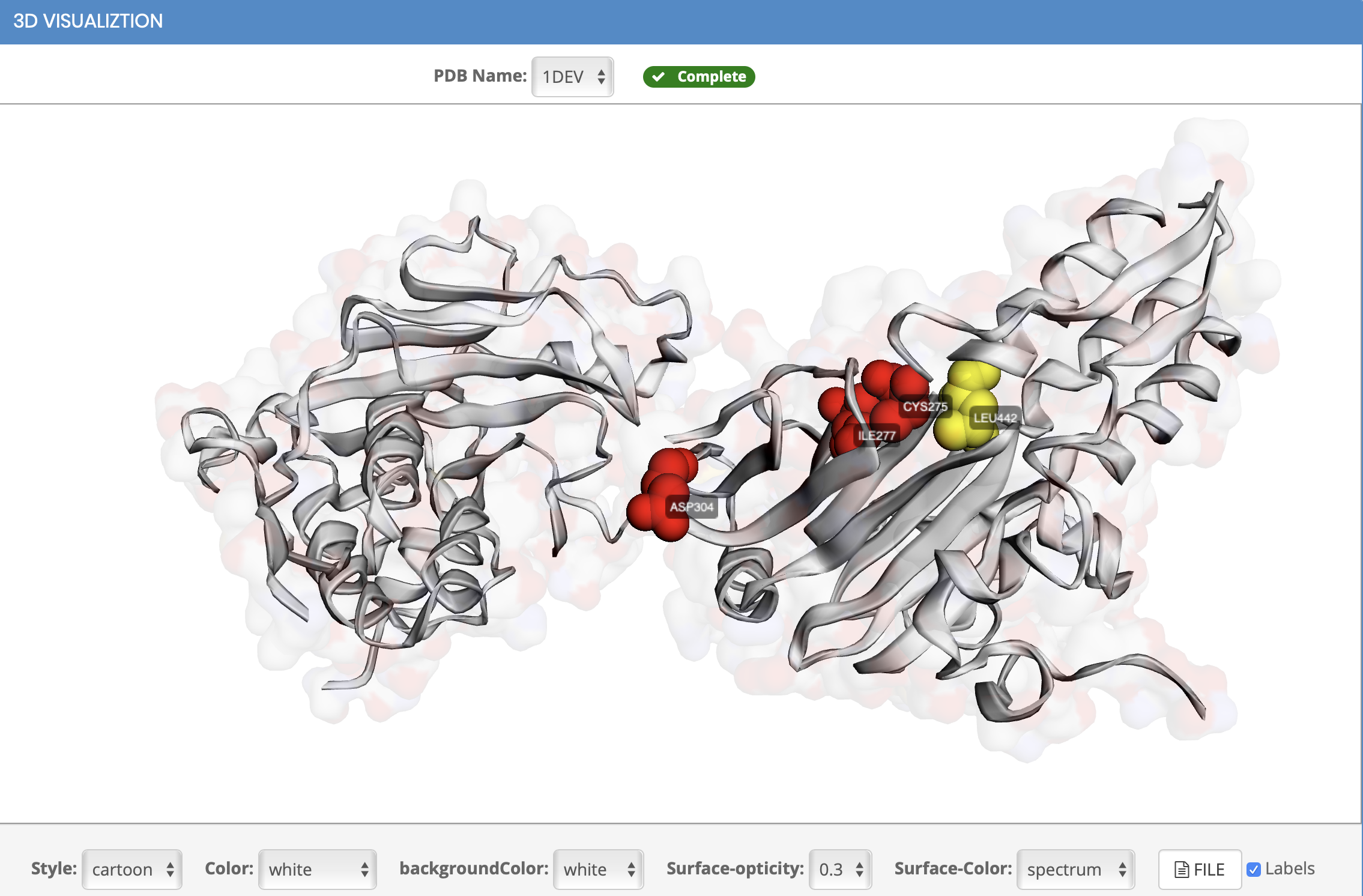
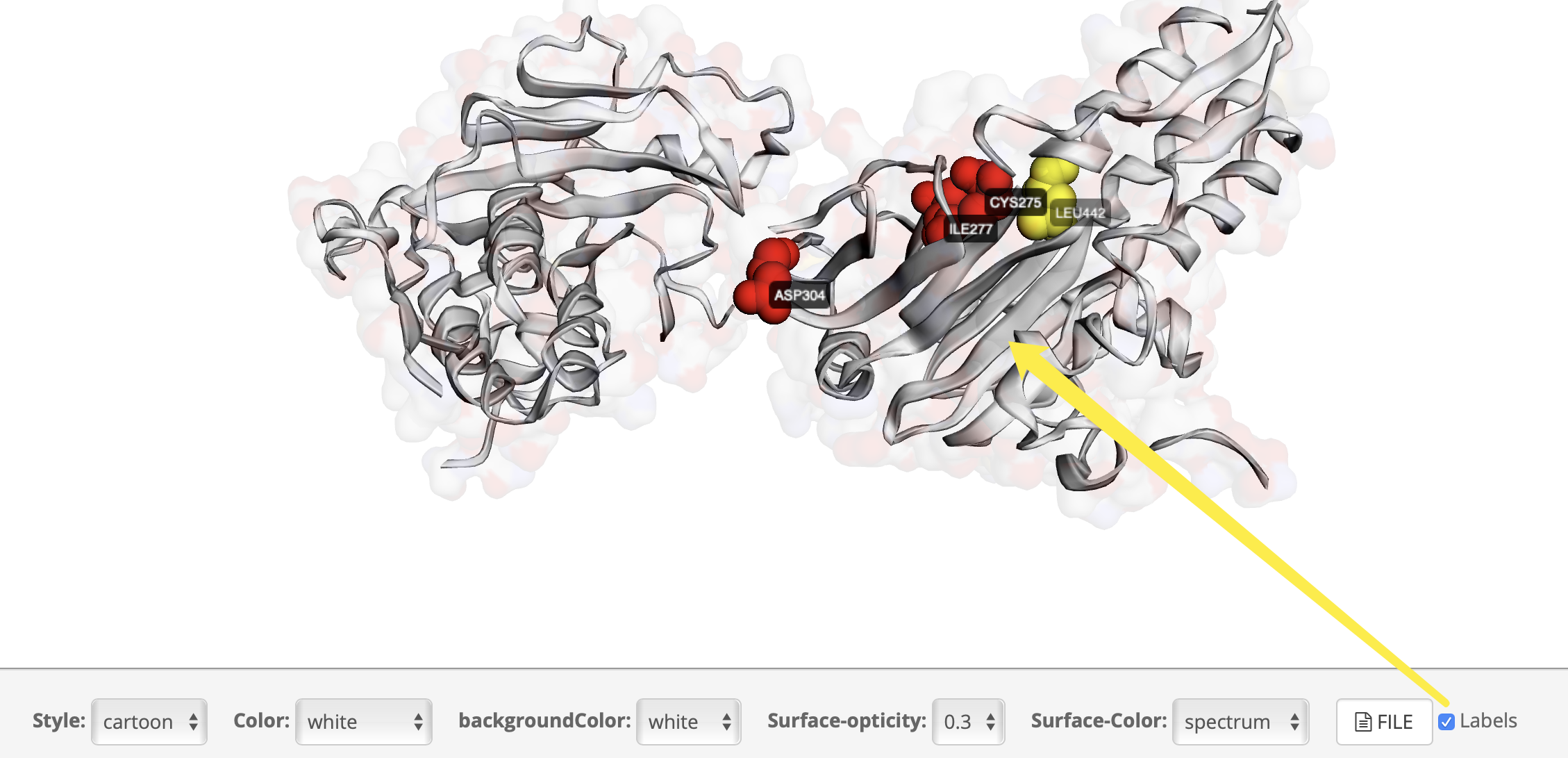
How to view the 3D structure?
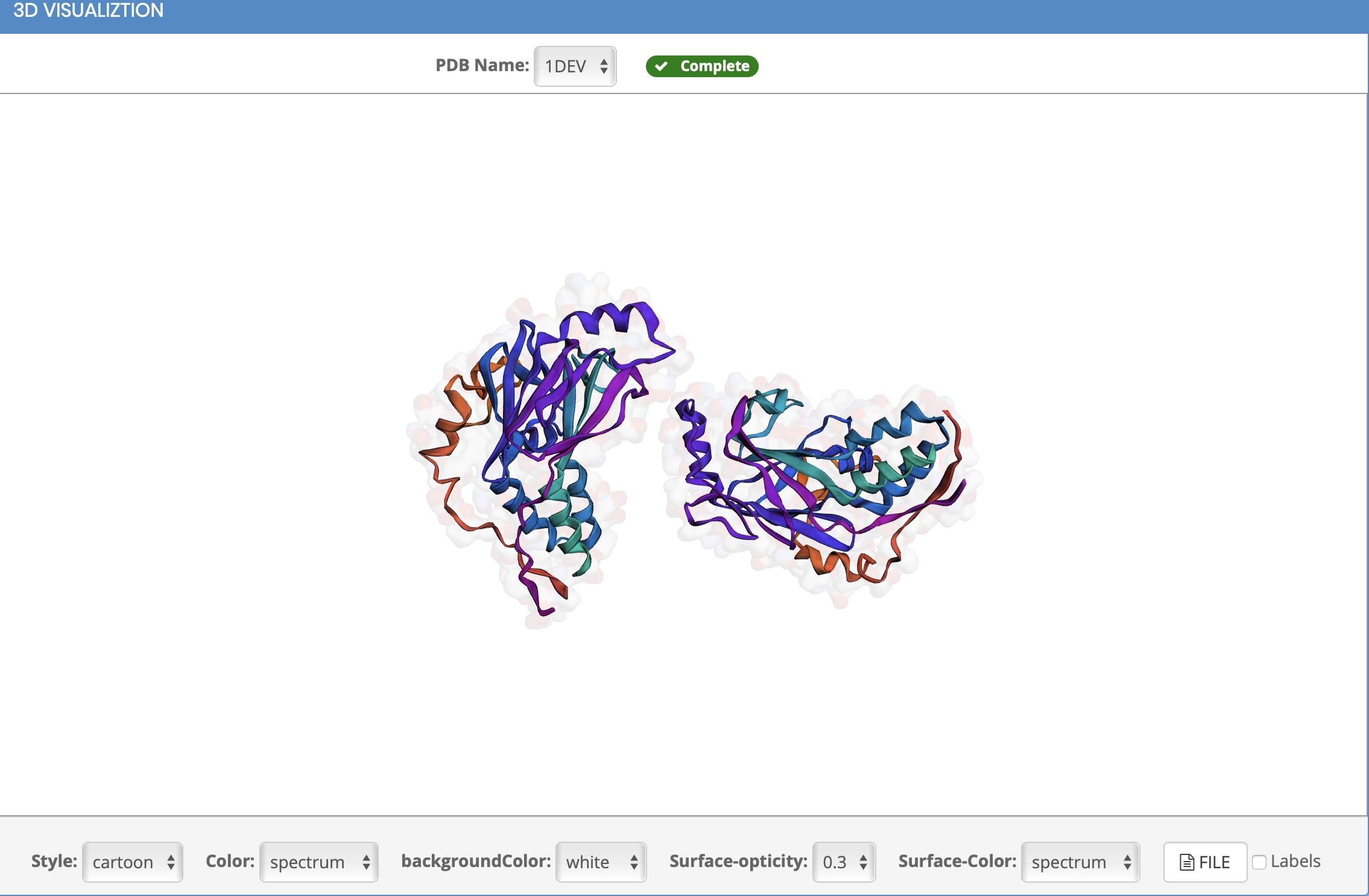
Can I customize the protein structure?
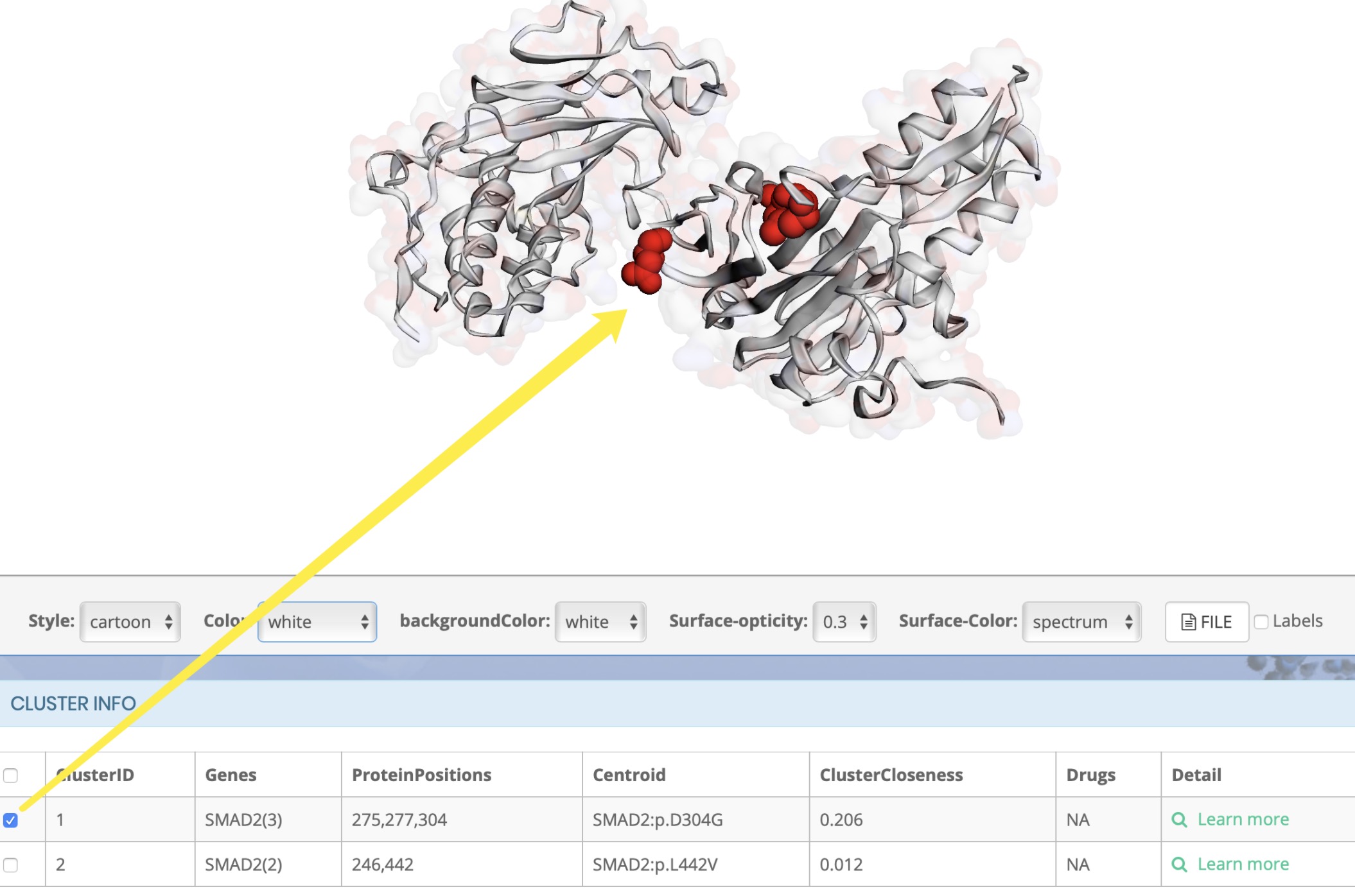
How to display cluster results
Why did the cluster appear on the protein structure after clicking?
How to distinguish cluster?
How to re-visualize?
HOW TO GET SUMMARY AND PAIRWISE FILE ?
COULD I RE-VISULATION AFTER 24 HOURS ?
WHEN SHOULD I USE RE-VISISUALATION ?
What about Result ?
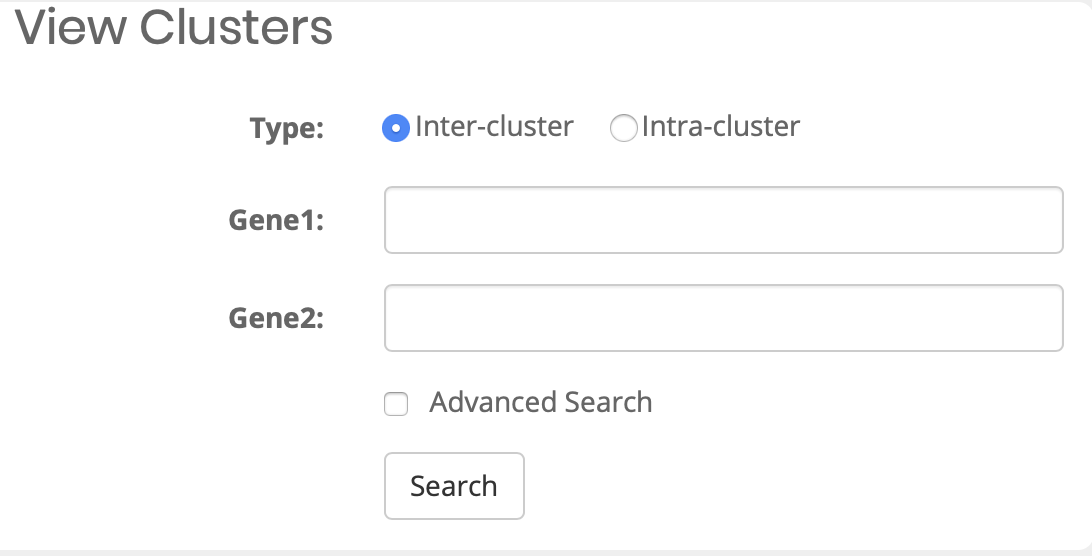
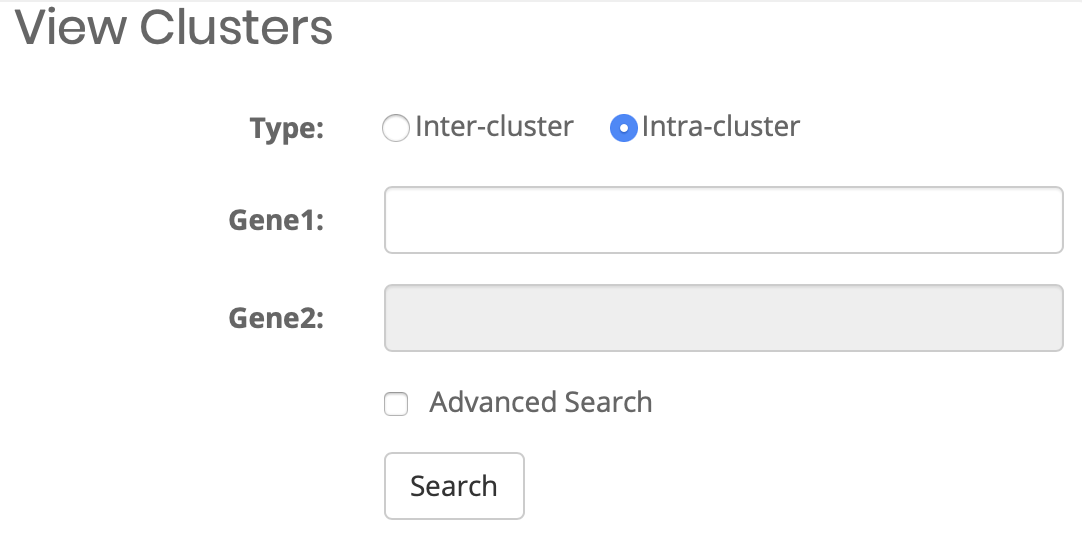
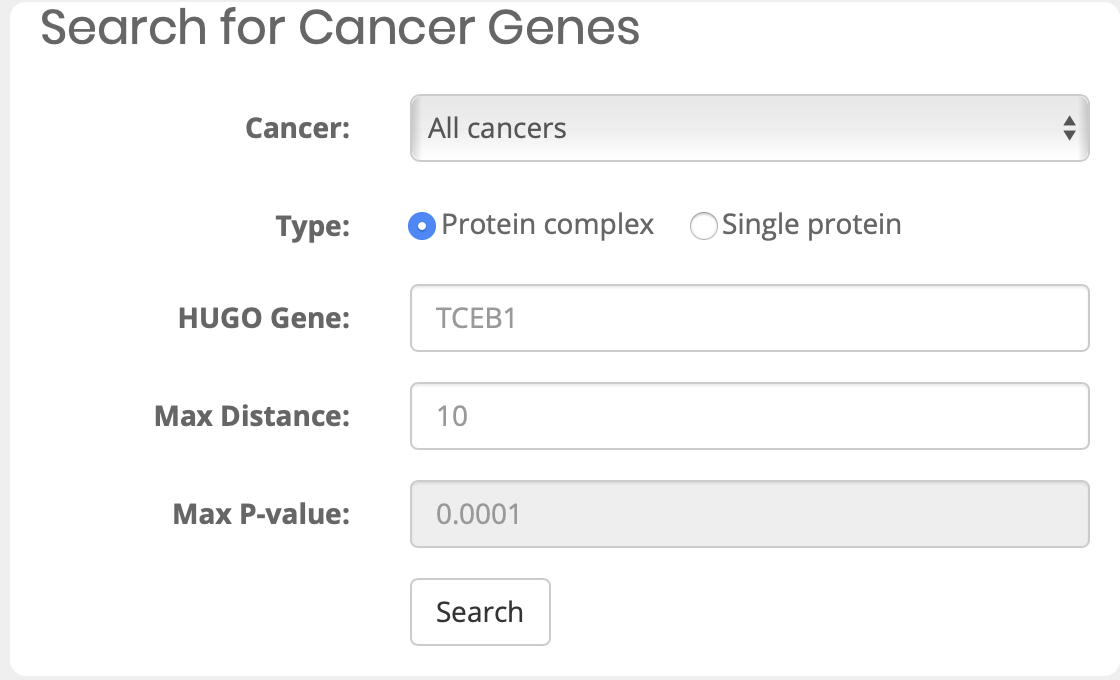
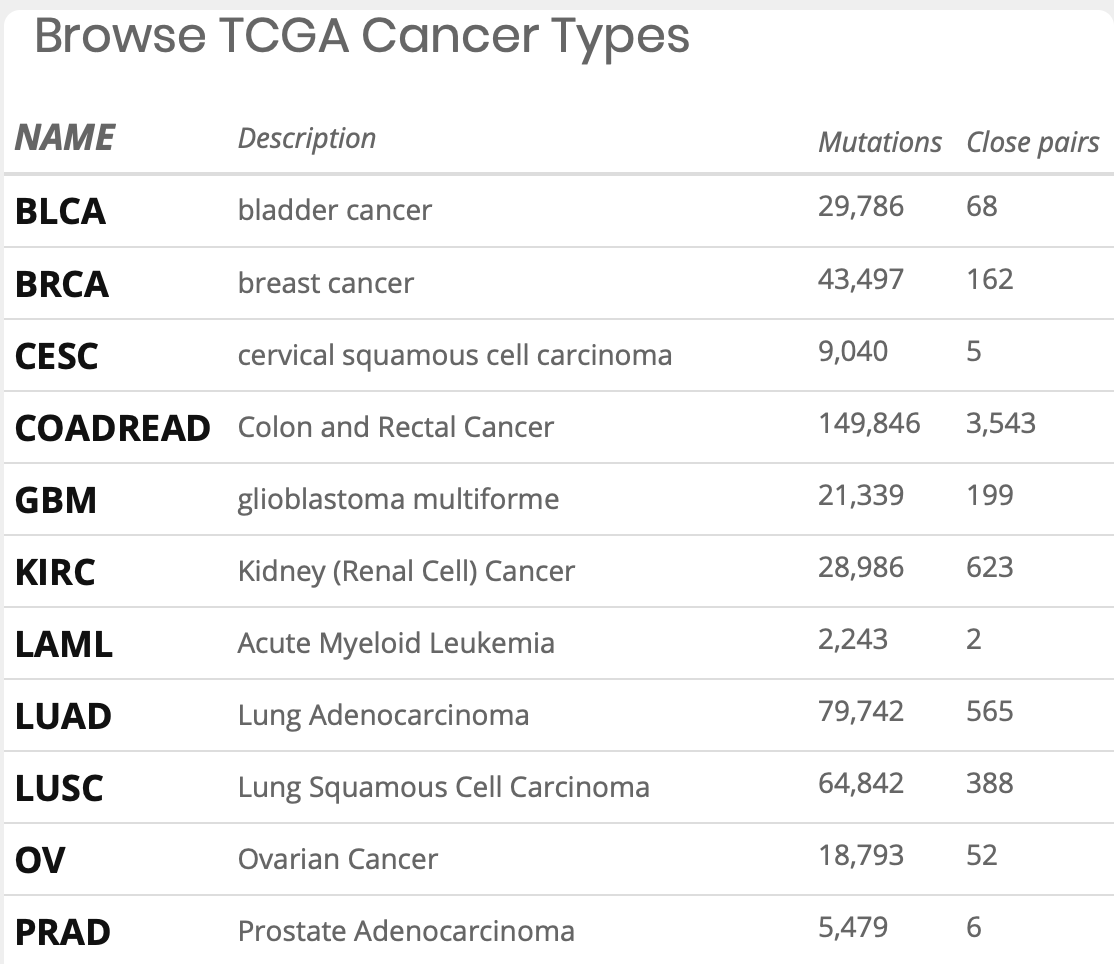
What can I query?
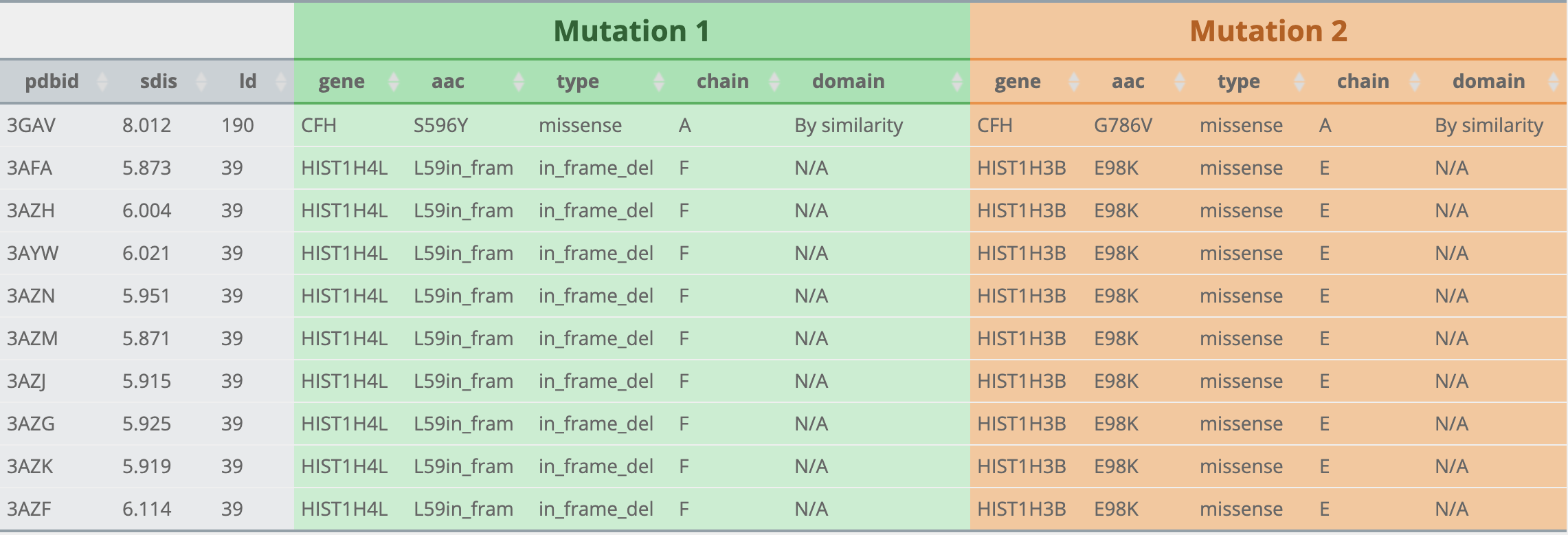
What is Mutation1 and Mutation2 after choosing cancer type?
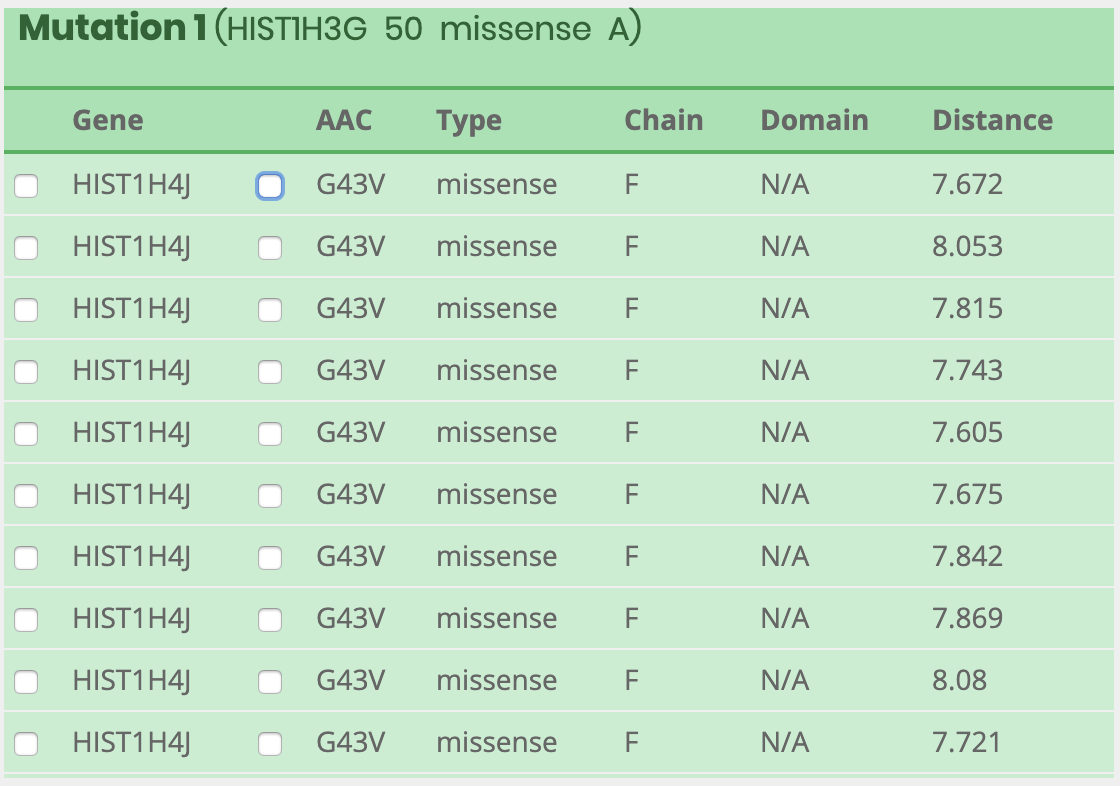
How to visualize after searching results?
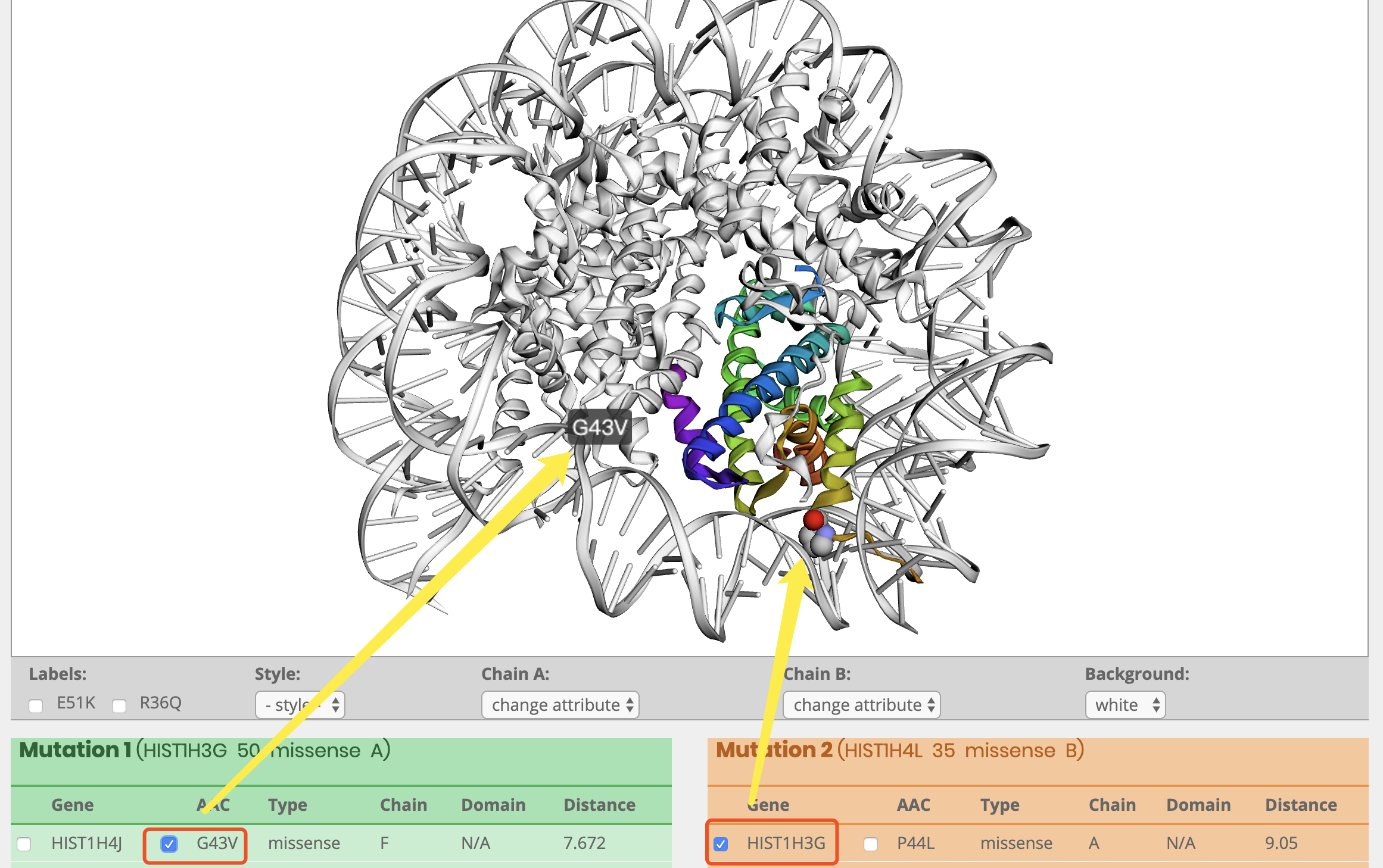
What information is included in the visual panel?
How to display clsuter and labels?
If you use HotSpot3D in your research, please quote
COMMERCIAL